By accessing this data, you agree to the terms of the MouseBIRN Data Use Agreement.
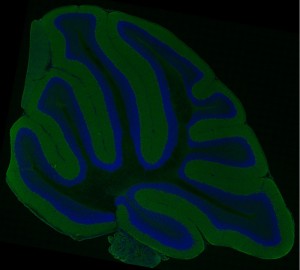
These data provide a high resolution, large scale mosaic image of a cerebellar distribution of cell bodies and alpha-synuclein from a non-transgenic animal (M. Martone; UCSD).
Description
A high resolution multiphoton microscopy mosaic image of a cerebellar distribution of cell bodies (Hoescht 33342; blue) and alpha-synuclein (green) from a non-transgenic animal.
This large scale image was acquired using a customized RTS2000 multi-photon microscope (Fan et al., 1999) equipped with a custom automated high precision motorized stage (Applied Precision LLC, Issaquah, WA, USA), which allows for the automatic acquisition of ultra-large field image mosaics in 2 and 3 dimensions (Price et al., In Press; Chow et al., Submitted). A Nikon Plan Apo TIRF (60X 1.45) oil immersion objective was used. These mosaic images are acquired by rastering the specimen along the X, Y, and Z axes, introducing a prescribed amount of overlap between acquired images (in this case 10%) to aid alignment. Unprocessed data acquired on the RTS2000 microscope is subsequently stored as a single stack of images. The image stack is analyzed using the JAVA-based ImageJ, a freely available software package, using plugins developed at NCMIR for processing, aligning, and assembling these massive datasets. Briefly, each file is separated into three separate .tiff stacks, one for each channel. Each tile is normalized to eliminate shading gradients, followed by the automatic alignment of individual tiles to form a full size mosaic image of the data for each channel. A globally optimized, accuracy. The assembled mosaics are then combined into one full-scale color image. For 3D imaging, the process is repeated for each wide field image plane in Z. The resulting image mosaics provide detailed views of cellular and subcellular structure and macromolecular distributions in a larger tissue context. These maps are being used to characterize Parkinson’s Disease. These data will provide the bridge between whole brain imaging techniques such as MRI (Duke University, CIVM) and electron microscopic (UCSD-NCMIR) analysis.
Accession Number
TBD
Download
The individual data files for the cerebellum mosaic are listed below:
Images
- Thumbnail (20 KB) – Download
- Scaled JPG (192 KB) – Download
- Original JPG (46.6 MB) – Download
- Original TIFF (697 MB) – Download
Access via CCDB
This data can also be accessed via registration through the Cell Centered Database (ccdb.ucsd.edu). Upon login, the user may query the database using the Project ID number 1187. The dataset ID is 102003b. Raw (*.img) and processed data (*.tiff) are freely available, as well as accompanying specimen preparation details (*.pdf).
Citations to Include
D.L. Price, S.K. Chow, H. Hakozaki, V. Phung, B. Smarr, S. Peltier, M.E. Martone, and M.H. Ellisman Application of a Multi-Photon High-Resolution Large-Scale Montage Imaging Technique to Characterize Transgenic Mouse Models of Human Neurodisorders, 2003 Microscopy & Microanalysis conference proceedings, Savannah, GA.
Price DL, Chow SK, MacLean NAB, Hakozaki H, Peltier S, Martone ME, Ellisman MH (2006) High-Resolution Large-Scale Mosaic Imaging using Multiphoton Microscopy to Characterize Transgenic Mouse Models of Human Neurological Disorders. Neuroinformatics 4(1):65-80
Contributors
National Center for Microscopy and Imaging Research at UC San Diego
Diana Price, Ph.D
Mark H. Ellisman, Ph.D.
Maryann Martone, Ph.D.
Technical Contact
Diana L. Price, Ph.D. (diana *AT* ncmir.ucsd.edu)
Acknowledgements
This work was supported by The Branfman Family and MJ Fox Foundations, NCRR RR04050, NIDCD DC03192 (CCDB), RR043050 (Mouse BIRN), and NIH LM 07292.